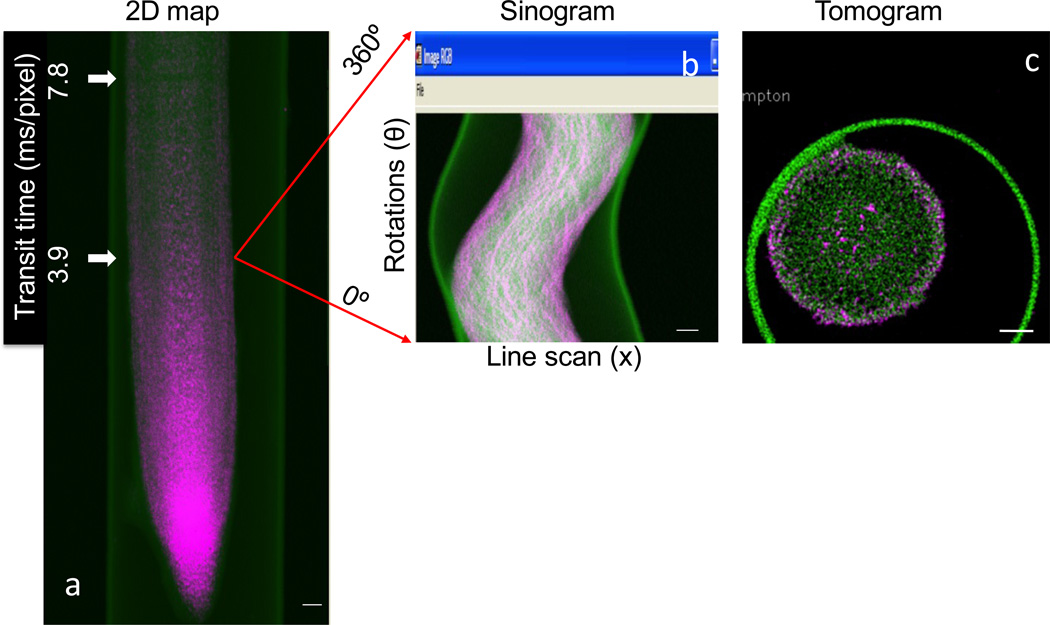
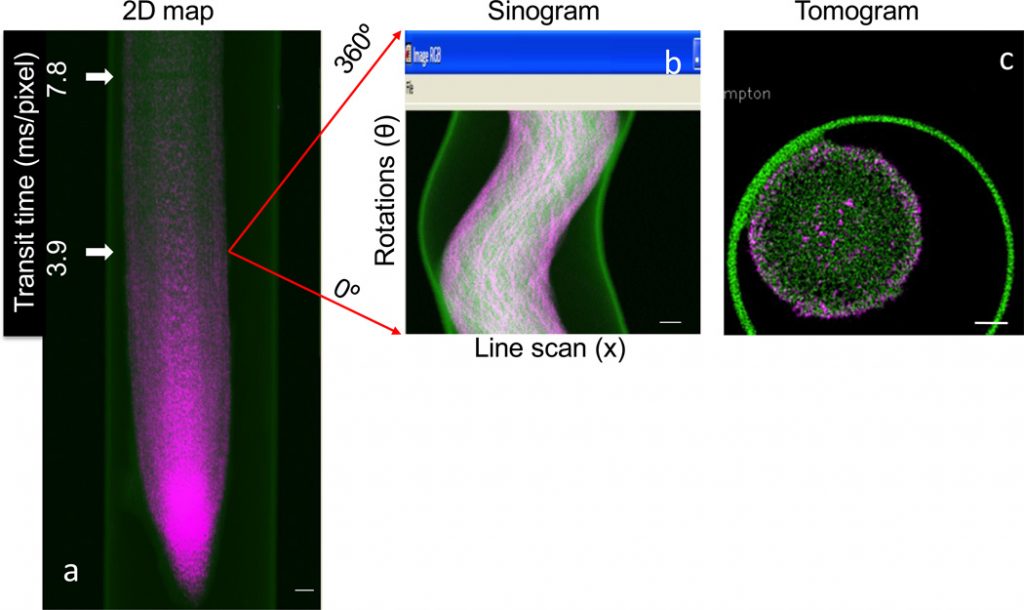
Functional characterisation of the genes regulating steel(loid) homeostasis in vegetation is a significant focus for phytoremediation, crop biofortification and meals safety analysis. Recent advances in X-ray focussing optics and fluorescence detection have tremendously improved the potential to make use of synchrotron techniques in plant science analysis.
With use of strategies resembling micro X-ray fluorescence mapping, micro computed tomography and micro X-ray absorption close to edge spectroscopy, steel(loids) might be imaged in vivo in hydrated plant tissues at submicron decision, and laterally resolved steel(loid) speciation can be decided beneath physiologically related circumstances.
This article focuses on the advantages of combining molecular biology and synchrotron-based techniques. By utilizing molecular techniques to probe the location of gene expression and protein manufacturing in mixture with laterally resolved synchrotron techniques, one can successfully and effectively assign practical info to particular genes.
A evaluation of the state of the artwork in this area is introduced, along with examples as to how synchrotron-based strategies might be mixed with molecular techniques to facilitate practical characterisation of genes in planta.
The article concludes with a abstract of the technical challenges nonetheless remaining for synchrotron-based laborious X-ray plant science analysis, notably these referring to subcellular stage analysis.
Population biology of fungal plant pathogens.
Studies of the inhabitants genetics of fungal and oomycetous phytopathogens are important to clarifying the illness epidemiology and devising administration methods.
Factors generally related to larger organisms resembling migration, pure choice, or recombination, are essential for the constructing of a clearer image of the pathogen in the panorama. In this chapter, we give attention to a restricted quantity of experimental and analytical strategies which are generally utilized in inhabitants genetics.
At first, we current differing kinds of qualitative and quantitative traits that might be recognized morphologically (phenotype). Subsequently, we describe a number of molecular strategies primarily based on dominant and codominant markers, and we offer our evaluation of the benefits and shortfalls of these strategies.
Third, we focus on numerous analytical strategies, which embody phylogenies, abstract statistics in addition to coalescent-based strategies, and we elaborate on the advantages related to every strategy. Last, we develop a case research in which we examine the inhabitants construction of the fungal phytopathogen Verticillium dahliae in coastal California, and assess the hypotheses of transcontinental gene move and recombination in a fungus that’s described as asexual.